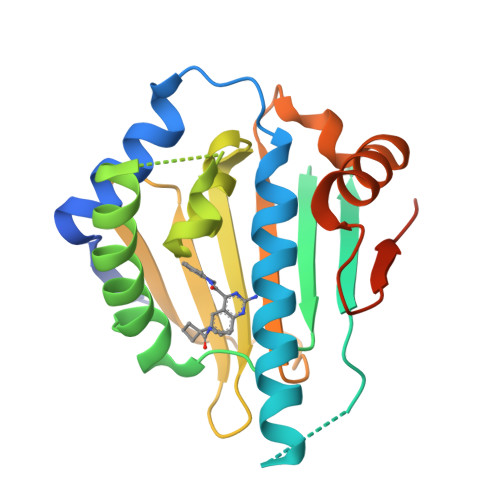
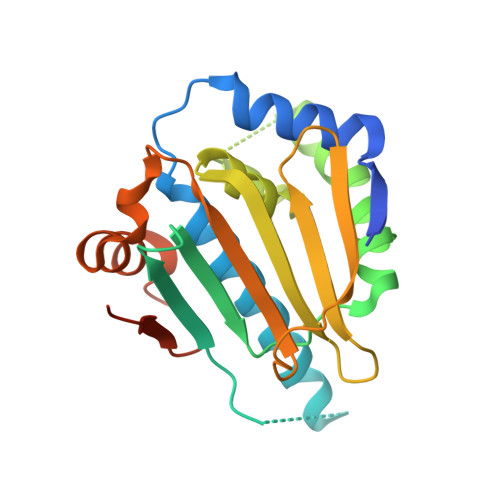
Ligand Desolvation Steers On-Rate and Impacts Drug Residence Time of Heat Shock Protein 90 (Hsp90) Inhibitors.
Schuetz, D.A., Richter, L., Amaral, M., Grandits, M., Gradler, U., Musil, D., Buchstaller, H.P., Eggenweiler, H.M., Frech, M., Ecker, G.F.(2018) J Med Chem 61: 4397-4411
- PubMed: 29701469
- DOI: https://doi.org/10.1021/acs.jmedchem.8b00080
- Primary Citation of Related Structures:
5LNY, 5LNZ, 5LO0, 5LO1, 5NYH, 5OCI, 5OD7, 5ODX - PubMed Abstract:
Residence time and more recently the association rate constant k on are increasingly acknowledged as important parameters for in vivo efficacy and safety of drugs. However, their broader consideration in drug development is limited by a lack of knowledge of how to optimize these parameters. In this study on a set of 176 heat shock protein 90 inhibitors, structure-kinetic relationships, X-ray crystallography, and molecular dynamics simulations were combined to retrieve a concrete scheme of how to rationally slow down on-rates. We discovered that an increased ligand desolvation barrier by introducing polar substituents resulted in a significant k on decrease. The slowdown was accomplished by introducing polar moieties to those parts of the ligand that point toward a hydrophobic cavity. We validated this scheme by increasing polarity of three Hsp90 inhibitors and observed a 9-, 13-, and 45-fold slowdown of on-rates and a 9-fold prolongation in residence time. This prolongation was driven by transition state destabilization rather than ground state stabilization.
Organizational Affiliation:
Department of Pharmaceutical Chemistry , University of Vienna , UZA 2, Althanstrasse 14 , 1090 Vienna , Austria.